Différences entre les versions de « Non covalent Interactions »
| (6 versions intermédiaires par le même utilisateur non affichées) | |||
| Ligne 1 : | Ligne 1 : | ||
[[Topological_Approaches_to_Intermolecular_Interactions|<<< Topological Approaches to Intermolecular Interactions workshop main page]] | [[Topological_Approaches_to_Intermolecular_Interactions|<<< Topological Approaches to Intermolecular Interactions workshop main page]] | ||
| − | === | + | === Introduction === |
| + | |||
Revealing the chemical bonding (structure) and the reorganization of the chemical bonds (reactivity) of any molecular system forms the undisputed foundation of chemistry. Chemical interactions between a protein and a drug, or a catalyst and its substrate, self-assembly of nanomaterials, and also many chemical reactions are dominated by noncovalent interactions. This class of interactions spans a wide range of binding energies and encompasses hydrogen bonding, dipole−dipole interactions, steric repulsion, and London dispersion. Molecular structure is governed by covalent, noncovalent, and electrostatic interactions, the latter two of which are the driving force in most biochemical processes. | Revealing the chemical bonding (structure) and the reorganization of the chemical bonds (reactivity) of any molecular system forms the undisputed foundation of chemistry. Chemical interactions between a protein and a drug, or a catalyst and its substrate, self-assembly of nanomaterials, and also many chemical reactions are dominated by noncovalent interactions. This class of interactions spans a wide range of binding energies and encompasses hydrogen bonding, dipole−dipole interactions, steric repulsion, and London dispersion. Molecular structure is governed by covalent, noncovalent, and electrostatic interactions, the latter two of which are the driving force in most biochemical processes. | ||
| + | {| border="0" | ||
| + | | | ||
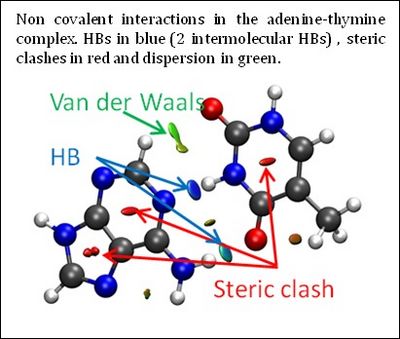
| + | The three-dimensional molecular structure defines covalent bonds; however, noncovalent interactions are hidden within voids in the bonding network. Although there are several ways to view and analyze covalent and electrostatic interactions, analogously simple methods for noncovalent interactions are rare. The development of such methods aids understanding of the complex interactions between biomolecules and the design of self-assembled materials and drugs, among others. | ||
| − | |||
| − | |||
| − | |||
One such proposal is the noncovalent interaction (NCI) index. It enables visualization and quantification of noncovalent theoretical results. NCI enables identification of interactions in 3D space from the electron density. Moreover, this approach has the advantage of being transferable to large systems, while remaining very fast to compute. Consequently, a qualitative NCI analysis is applicable to extremely large systems, including, e.g. proteins and DNA, where describing the interplay of attractive and repulsive interactions is crucial for understanding functionality. This new index, requiring only molecular geometry information, compliments existing methods for covalent and electrostatic interactions, providing a perfect bridge between theoretical chemistry and (in)organic chemistry by means of chemical bonds and their change. | One such proposal is the noncovalent interaction (NCI) index. It enables visualization and quantification of noncovalent theoretical results. NCI enables identification of interactions in 3D space from the electron density. Moreover, this approach has the advantage of being transferable to large systems, while remaining very fast to compute. Consequently, a qualitative NCI analysis is applicable to extremely large systems, including, e.g. proteins and DNA, where describing the interplay of attractive and repulsive interactions is crucial for understanding functionality. This new index, requiring only molecular geometry information, compliments existing methods for covalent and electrostatic interactions, providing a perfect bridge between theoretical chemistry and (in)organic chemistry by means of chemical bonds and their change. | ||
| + | | | ||
| + | [[File:AT.jpg|400px]] | ||
| + | |} | ||
Dernière version du 9 janvier 2013 à 10:22
<<< Topological Approaches to Intermolecular Interactions workshop main page
Introduction
Revealing the chemical bonding (structure) and the reorganization of the chemical bonds (reactivity) of any molecular system forms the undisputed foundation of chemistry. Chemical interactions between a protein and a drug, or a catalyst and its substrate, self-assembly of nanomaterials, and also many chemical reactions are dominated by noncovalent interactions. This class of interactions spans a wide range of binding energies and encompasses hydrogen bonding, dipole−dipole interactions, steric repulsion, and London dispersion. Molecular structure is governed by covalent, noncovalent, and electrostatic interactions, the latter two of which are the driving force in most biochemical processes.
|
The three-dimensional molecular structure defines covalent bonds; however, noncovalent interactions are hidden within voids in the bonding network. Although there are several ways to view and analyze covalent and electrostatic interactions, analogously simple methods for noncovalent interactions are rare. The development of such methods aids understanding of the complex interactions between biomolecules and the design of self-assembled materials and drugs, among others.
|