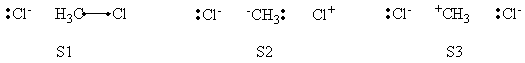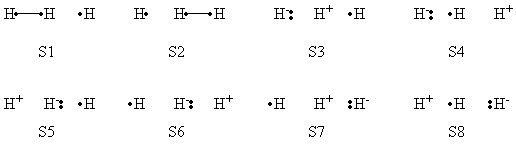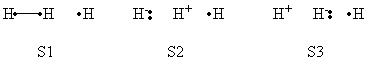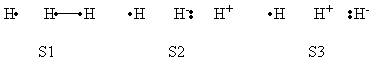Différences entre les versions de « VBTutorial3 »
| (38 versions intermédiaires par 5 utilisateurs non affichées) | |||
| Ligne 1 : | Ligne 1 : | ||
| − | [[ | + | [[VB_tutorial|<<< VB tutorials main page]] |
| Ligne 6 : | Ligne 6 : | ||
== Exercise 1 : Computation of state correlation Diagrams for a 3 centers / 4 electrons system == | == Exercise 1 : Computation of state correlation Diagrams for a 3 centers / 4 electrons system == | ||
| − | In this exercise the <math>\textrm{S}_{\textrm{N}}2</math> reaction Cl<math>{}^{-}</math> + | + | In this exercise the <math>\textrm{S}_{\textrm{N}}2</math> reaction Cl<math>{}^{-}</math> + CH<math>{}_3</math>Cl -> ClCH<math>{}_3</math> + Cl<math>{}^{-}</math> will be studied in both vacuum and solution. Valence Bond State Correlation Diagrams (VBSCD) will be constructed at <math>\pi</math>-D-BOVB level. There are two parts in this exercise: basic part and optional part. The basic part is performed with MCP-DZP basis set in which the inner orbitals in Cl and C are described with MCP pseudo potential. The optional part is performed with 6-31+G* basis set, using the general specification for the xmvb input (expert users). Only reactant and transition state will be computed in this exercise, which is sufficient to build the VBSCD diagrams. <br> |
{| class="collapsible collapsed wikitable" | {| class="collapsible collapsed wikitable" | ||
| Ligne 24 : | Ligne 24 : | ||
| | | | ||
==== 1. Compute the Energies and Wavefunctions at Reactant and Transition State with Different Sets of VB Structures==== | ==== 1. Compute the Energies and Wavefunctions at Reactant and Transition State with Different Sets of VB Structures==== | ||
| + | |||
| + | We advise you to create the first xmvb input file (''.xmi'' file) for this study starting from a ''.xmi'' file taken from tutorial1 as a template. Alternatively, you may copy the input file corresponding for the first calculation of this study (L-VBSCF on reactant state geometry in vacuum) from the ''answer'' folder of this exercise (''cp answer/rs_vac_vbscf.xmi .''), run directly the first calculation, inspect input/outputs, and then use this ''.xmi'' file as a template for the following calculations. | ||
| + | |||
# Write the following VB structure sets for a 3 centers / 4 electrons system : | # Write the following VB structure sets for a 3 centers / 4 electrons system : | ||
## all structures; | ## all structures; | ||
## minimal structures for reactant; | ## minimal structures for reactant; | ||
## minimal structures for product. | ## minimal structures for product. | ||
| + | # How would you define your different fragment orbitals for this calculation (''$frag'' section) ? You will find the answer for this case is in the [[General_guidelines_for_BOVB_calculations#Recommended_definition_for_the_orbital_blocks|"recommended definition for the orbital blocks" section of the "practical for BOVB calculations" document]] | ||
# Perform <math>\pi</math>-D-BOVB calculation for reactant [[General_guidelines_for_BOVB_calculations#High_symmetry_case:|(see "high symmetry cases" here)]]: | # Perform <math>\pi</math>-D-BOVB calculation for reactant [[General_guidelines_for_BOVB_calculations#High_symmetry_case:|(see "high symmetry cases" here)]]: | ||
## Perform all-structure <math>\pi</math>-D-BOVB calculation as following: | ## Perform all-structure <math>\pi</math>-D-BOVB calculation as following: | ||
| − | ### Perform L-VBSCF calculation with "''orbtyp=hao frgtyp=sao guess=mo''", in which the orbitals are all localized on the Cl and CH<math>{}_3</math> groups; | + | ### Perform L-VBSCF (file name: rs_vac_vbscf) calculation with "''orbtyp=hao frgtyp=sao guess=mo''", in which the orbitals are all localized on the Cl and CH<math>{}_3</math> groups; |
| − | ### Perform π-D-VBSCF calculation where <math>\pi</math> orbitals are delocalized in the whole system and the <math>\sigma</math> orbitals are kept localized. Use the L-VBSCF orbitals as initial guess; | + | ### Perform π-D-VBSCF calculation where <math>\pi</math> orbitals are delocalized in the whole system and the <math>\sigma</math> orbitals are kept localized (file name: rs_vac_d-vbscf). Use the L-VBSCF orbitals as initial guess; |
| − | ### Perform π-D-BOVB calculation with π-D-VBSCF orbitals as initial guess. | + | ### Perform π-D-BOVB (file name: rs_vac_d-bovb) calculation with π-D-VBSCF orbitals as initial guess. |
| − | ## Perform <math>\pi</math>-D-BOVB calculations with minimal structures for reactant and product with the same procedure as all-structure calculation. | + | ## Perform <math>\pi</math>-D-BOVB calculations with minimal structures for reactant (file names: rs_vac_*_rs) and product (file names: rs_vac_*_ps) with the same procedure as all-structure calculation. |
| − | # Perform <math>\pi</math>-D-BOVB calculation for transition state. The procedure is the same as step 2. | + | # Perform <math>\pi</math>-D-BOVB calculation for transition state (file names: ts_vac_*). The procedure is the same as step 2. |
# Perform <math>\pi</math>-D-BOVB/PCM calculations for reactant: | # Perform <math>\pi</math>-D-BOVB/PCM calculations for reactant: | ||
| − | ## Perform all-structure <math>\pi</math>-D-BOVB/PCM calculation for the reactant | + | ## Perform all-structure <math>\pi</math>-D-BOVB/PCM (file name: rs_pcm_d-bovb) calculation for the reactant, starting directly from the <math>\pi</math>-D-BOVB orbitals computed in vacuum (2.1) |
| − | + | ## Perform <math>\pi</math>-D-BOVB/PCM calculations with minimal structures for reactant (file name: rs_pcm_d-bovb_rs) and product (file name: rs_pcm_d-bovb), also starting from the corresponding <math>\pi</math>-D-BOVB orbitals computed in vacuum (2.2) | |
| − | + | # Perform BOVB/PCM calculations for transition state (file names: rs_pcm_d-bovb*) with the same procedure as step 4. | |
| − | |||
| − | ## Perform <math>\pi</math>-D-BOVB/PCM calculations with minimal structures for reactant and product in | ||
| − | # Perform BOVB/PCM calculations for transition state with the same procedure as step 4. | ||
==== 2. Analysis: Wavefunctions and Energies==== | ==== 2. Analysis: Wavefunctions and Energies==== | ||
| Ligne 47 : | Ligne 48 : | ||
# Compute the Barrier height of the <math>\textrm{S}_{\textrm{N}}2</math> reaction in both vacuum and solution. See the difference of the barrier heights, and find out the reason. | # Compute the Barrier height of the <math>\textrm{S}_{\textrm{N}}2</math> reaction in both vacuum and solution. See the difference of the barrier heights, and find out the reason. | ||
# Compare the energies of reactant and product structures at reactant and transition state geometries, in both vacuum and solution. What's the difference of the energies at different points? Why? | # Compare the energies of reactant and product structures at reactant and transition state geometries, in both vacuum and solution. What's the difference of the energies at different points? Why? | ||
| − | # Compute the resonance | + | # Compute the resonance energy at the crossing point of diabatic curves state points in vacuum. |
{| class="collapsible collapsed wikitable" | {| class="collapsible collapsed wikitable" | ||
| Ligne 63 : | Ligne 64 : | ||
<center>[[File:ClCH3Cl_Product_Structures.png|600px]]</center><br> | <center>[[File:ClCH3Cl_Product_Structures.png|600px]]</center><br> | ||
<center>VB Structure Set of The Product</center> | <center>VB Structure Set of The Product</center> | ||
| − | |||
| − | |||
| − | |||
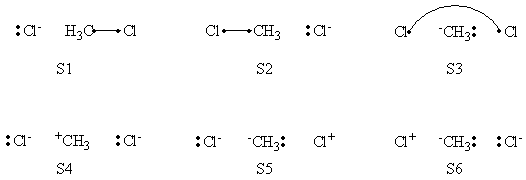
=====Weights of Structures===== | =====Weights of Structures===== | ||
| Ligne 83 : | Ligne 81 : | ||
|- | |- | ||
! scope="row" | BOVB | ! scope="row" | BOVB | ||
| − | |0. | + | |0.587 || || || 0.352 || 0.061 || |
|- | |- | ||
! scope="row" | VBSCF/PCM | ! scope="row" | VBSCF/PCM | ||
| Ligne 89 : | Ligne 87 : | ||
|- | |- | ||
! scope="row" | BOVB/PCM | ! scope="row" | BOVB/PCM | ||
| − | |0. | + | |0.639 || || || 0.413 || 0.131 || |
|} | |} | ||
</center> | </center> | ||
| Ligne 108 : | Ligne 106 : | ||
|- | |- | ||
! scope="row" | BOVB | ! scope="row" | BOVB | ||
| − | | 0.228 || 0.228 || 0.041 || 0. | + | | 0.228 || 0.228 || 0.041 || 0.502 || || |
|- | |- | ||
! scope="row" | VBSCF/PCM | ! scope="row" | VBSCF/PCM | ||
| Ligne 114 : | Ligne 112 : | ||
|- | |- | ||
! scope="row" | BOVB/PCM | ! scope="row" | BOVB/PCM | ||
| − | |0.215 || 0.215 || 0. | + | |0.215 || 0.215 || 0.034 || 0.536 || || |
|} | |} | ||
</center> | </center> | ||
| Ligne 129 : | Ligne 127 : | ||
|- | |- | ||
! scope="row" |Reactant | ! scope="row" |Reactant | ||
| − | | -37.03384 || -37. | + | | -37.03384 || -37.05085 || -37.13644 || -37.15356 |
|- | |- | ||
! scope="row" | Trasition State | ! scope="row" | Trasition State | ||
| − | | -36.98034 || -37. | + | | -36.98034 || -37.02508 || -37.06980 || -37.11346 |
|- | |- | ||
! scope="row" | Barrier | ! scope="row" | Barrier | ||
| − | | 33.6 || 16. | + | | 33.6 || 16.2 || 41.8 || 25.2 |
|} | |} | ||
</center> | </center> | ||
| Ligne 146 : | Ligne 144 : | ||
! scope="col" | VBSCF | ! scope="col" | VBSCF | ||
! scope="col" | BOVB | ! scope="col" | BOVB | ||
| − | |||
! scope="col" | BOVB/PCM | ! scope="col" | BOVB/PCM | ||
|- | |- | ||
! scope="row" | All Structures | ! scope="row" | All Structures | ||
| − | | -36.98034 ||-37. | + | | -36.98034 ||-37.02508 || -37.11346 |
|- | |- | ||
! scope="row" | Reactant | ! scope="row" | Reactant | ||
| − | | -36. | + | | -36.95984 || -36.98719 || -37.07951 |
|- | |- | ||
! scope="row" | Product | ! scope="row" | Product | ||
| − | | -36. | + | | -36.95984 ||-36.98718 || -37.07949 |
|- | |- | ||
! scope="row" | Resonance Energy | ! scope="row" | Resonance Energy | ||
| − | | | + | | 12.9 || 23.8 || 21.3 |
| + | |} | ||
| + | </center> | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | {| class="collapsible collapsed wikitable" | ||
| + | |- | ||
| + | !<big>'''Optional part (expert users)'''</big> | ||
| + | |- | ||
| + | | | ||
| + | In this part, calculations with BFI section are performed, which is a technique for the experienced users. The 6-31+G* basis set is used. The inner orbitals are frozen as HF orbitals in all VB calculations and the valence basis functions are reorganized to hybrid basis functions so that the <math>\sigma</math>, <math>\pi_x</math> and <math>\pi_y</math> spaces can be separated well. A D-BOVB calculation is performed in 2 steps: | ||
| + | # Perform a VBSCF calculation with <math>\pi</math> orbitals delocalized in the whole system and <math>\sigma</math> orbitals localized on the Cl and CH<math>{}_3</math> groups; | ||
| + | # Perform a BOVB calculation with VBSCF orbitals as initial guess. | ||
| + | The VB calculations are the same as the calculations performed above. Try to understand the BFI section, perform the calculations and compare the differences of barrier heights, resonance energies and cpu performances with and without $BFI. | ||
| + | |||
| + | {| class="collapsible collapsed wikitable" | ||
| + | |- | ||
| + | !'''Answer''' | ||
| + | |- | ||
| + | | | ||
| + | |||
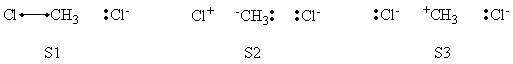
| + | =====Weights of Structures===== | ||
| + | <center> | ||
| + | {| border="1" | ||
| + | |+ Weights of structures at Reactant Geometry | ||
| + | ! scope="col" | | ||
| + | ! scope="col" | S1 | ||
| + | ! scope="col" | S2 | ||
| + | ! scope="col" | S3 | ||
| + | ! scope="col" | S4 | ||
| + | ! scope="col" | S5 | ||
| + | ! scope="col" | S6 | ||
| + | |- | ||
| + | ! scope="row" |VBSCF | ||
| + | | 0.632 || 0.003 || 0.001 || 0.299 || 0.064 || 0.000 | ||
| + | |- | ||
| + | ! scope="row" | BOVB | ||
| + | | 0.566 || 0.061 || 0.005 || 0.288 || 0.081 || 0.000 | ||
| + | |- | ||
| + | ! scope="row" | VBSCF/PCM | ||
| + | | 0.645 || 0.002 || 0.001 || 0.279 || 0.073 || 0.000 | ||
| + | |- | ||
| + | ! scope="row" | BOVB/PCM | ||
| + | | 0.599 || -0.019 || -0.007 || 0.337 || 0.089 || 0.000 | ||
| + | |} | ||
| + | </center> | ||
| + | <br> | ||
| + | <center> | ||
| + | {| border="1" | ||
| + | |+ Weights of structures at Transition State Geometry | ||
| + | ! scope="col" | | ||
| + | ! scope="col" | S1 | ||
| + | ! scope="col" | S2 | ||
| + | ! scope="col" | S3 | ||
| + | ! scope="col" | S4 | ||
| + | ! scope="col" | S5 | ||
| + | ! scope="col" | S6 | ||
| + | |- | ||
| + | ! scope="row" |VBSCF | ||
| + | | 0.239 || 0.240 || 0.027 || 0.496 || -0.001 || -0.001 | ||
| + | |- | ||
| + | ! scope="row" | BOVB | ||
| + | | 0.263 || 0.263 || 0.043 || 0.416 || 0.007 || 0.007 | ||
| + | |- | ||
| + | ! scope="row" | VBSCF/PCM | ||
| + | | 0.220 || 0.220 || 0.020 || 0.542 || -0.001 || -0.001 | ||
| + | |- | ||
| + | ! scope="row" | BOVB/PCM | ||
| + | | 0.251 || 0.251 || 0.036 || 0.452 || 0.005 || 0.005 | ||
| + | |} | ||
| + | </center> | ||
| + | |||
| + | =====Barrier of the Reaction===== | ||
| + | <center> | ||
| + | {| border="1" | ||
| + | |+ Energies (a.u.) and Barriers (kcal/mol) of <math>\textrm{S}_{\textrm{N}}2</math> Reaction | ||
| + | ! scope="col" | | ||
| + | ! scope="col" | VBSCF | ||
| + | ! scope="col" | BOVB | ||
| + | ! scope="col" | VBSCF/PCM | ||
| + | ! scope="col" | BOVB/PCM | ||
| + | |- | ||
| + | ! scope="row" |Reactant | ||
| + | | -958.65818 || -958.67599 || -958.75774 || -958.78203 | ||
| + | |- | ||
| + | ! scope="row" | Trasition State | ||
| + | | -958.62157 || -958.65321 || -958.71004 || -958.73960 | ||
| + | |- | ||
| + | ! scope="row" | Barrier | ||
| + | | 23.0 || 14.3 || 29.9 || 26.6 | ||
|} | |} | ||
</center> | </center> | ||
| Ligne 166 : | Ligne 254 : | ||
<center> | <center> | ||
{| border="1" | {| border="1" | ||
| − | |+ Energies(a.u.) and Resonance Energies (kcal/mol) of | + | |+ Energies (a.u.) and Resonance Energies (kcal/mol) of Transition State |
! scope="col" | | ! scope="col" | | ||
! scope="col" | VBSCF | ! scope="col" | VBSCF | ||
| Ligne 174 : | Ligne 262 : | ||
|- | |- | ||
! scope="row" | All Structures | ! scope="row" | All Structures | ||
| − | | - | + | | -958.62157 || -958.65321 || -958.71004 || -958.73960 |
|- | |- | ||
! scope="row" | Reactant | ! scope="row" | Reactant | ||
| − | | - | + | | -958.59188 || -958.60816 || -958.68547 || -958.70142 |
|- | |- | ||
! scope="row" | Product | ! scope="row" | Product | ||
| − | | - | + | | -958.59188 || -958.60816 || -958.68547 || -958.70142 |
|- | |- | ||
! scope="row" | Resonance Energy | ! scope="row" | Resonance Energy | ||
| − | | | + | | 18.6 || 28.3 || 15.4 || 23.9 |
|} | |} | ||
</center> | </center> | ||
| − | |||
| − | | | + | =====Resonance Energies of Reactant===== |
| − | + | <center> | |
| − | + | {| border="1" | |
| + | |+ Energies(a.u.) and Resonance Energies (kcal/mol) of Reactant | ||
| + | ! scope="col" | | ||
| + | ! scope="col" | VBSCF | ||
| + | ! scope="col" | BOVB | ||
| + | ! scope="col" | VBSCF/PCM | ||
| + | ! scope="col" | BOVB/PCM | ||
| + | |- | ||
| + | ! scope="row" | All Structures | ||
| + | | -958.65818 || -958.67599 || -958.75774 || -958.78203 | ||
| + | |- | ||
| + | ! scope="row" | Reactant | ||
| + | | -958.56752 || -958.67272 || -958.75734 || -958.77250 | ||
|- | |- | ||
| − | ! | + | ! scope="row" | Product |
| + | | -958.31845 || -958.31845 || -958.57917 || -958.58246 | ||
|- | |- | ||
| − | | | + | ! scope="row" | Resonance Energy |
| − | + | | 0.4 || 2.1 || 0.3 || 6.0 | |
| − | + | |} | |
| − | + | </center> | |
| − | + | |} | |
| − | |||
|} | |} | ||
| − | |||
| − | |||
| − | |||
[[General_guidelines_for_BOVB_calculations| >> general guidelines for BOVB calculations]] | [[General_guidelines_for_BOVB_calculations| >> general guidelines for BOVB calculations]] | ||
| Ligne 210 : | Ligne 306 : | ||
{| class="collapsible collapsed wikitable" | {| class="collapsible collapsed wikitable" | ||
|- | |- | ||
| − | !<big>'''Optional exercises - Homework'''</big> | + | !<big><big><big>'''Optional exercises - Homework'''</big></big></big> |
|- | |- | ||
| | | | ||
== Exercise 2 : computation of H—H + H. -> H. + H—H radical exchange VBSCD diagram == | == Exercise 2 : computation of H—H + H. -> H. + H—H radical exchange VBSCD diagram == | ||
| − | |||
===1/ Paper exercise :=== | ===1/ Paper exercise :=== | ||
| Ligne 244 : | Ligne 339 : | ||
c/ It is known that for strong binders, at any given bonding distance the singlet-triplet transition energy is larger than twice the bonding energy of the dimer at equilibrium distance, so that one can write the approximate expression <big> <math>\Delta E_{ST} </math>' <math> = 2 BDE </math></big>, where <big><math> BDE </math></big> is the bonding energy of the dimer at equilibrium distance. Using the latter expression, express the avoided crossing term <big><math> B </math></big> as a function of the bonding energy of <big><math> H_{2}</math></big>. | c/ It is known that for strong binders, at any given bonding distance the singlet-triplet transition energy is larger than twice the bonding energy of the dimer at equilibrium distance, so that one can write the approximate expression <big> <math>\Delta E_{ST} </math>' <math> = 2 BDE </math></big>, where <big><math> BDE </math></big> is the bonding energy of the dimer at equilibrium distance. Using the latter expression, express the avoided crossing term <big><math> B </math></big> as a function of the bonding energy of <big><math> H_{2}</math></big>. | ||
| + | {| class="collapsible collapsed wikitable" | ||
| + | |- | ||
| + | !'''Answer''' | ||
| + | |- | ||
| + | | | ||
| + | a - Considering the R and R* states for the radical exchange process, the HL wave functions for R and R* are the following :<br> | ||
| + | <math>\psi (R)=(|xa\overline{y}|-|x\overline{a}y|)(1/2)^{-1/2} </math><br> | ||
| + | <math>\psi (R^{*})=(|x\overline{a}y|-|\overline{x}ay|)(1/2)^{-1/2}</math><br> | ||
| + | At the equilibrium distance of R, A and Y are close together while X is far away. Therefore R is stabilized by a covalent bond and its energy is <big><math> 2 \beta S </math></big>. In R*, the AY entity has two parallel spins 50% of the time (a repulsive situation), and alternated spins 50% of the time (a non-bonding situation). Therefore the energy of R* is half a triplet repulsion, i.e. <big><math> - \beta S </math></big>. As <big><math>\Delta E_{ST} (A-Y)= 4 \beta S </math></big>, the final G expression is:<br> | ||
| + | <big><math>G=0.75 \Delta E_{ST} (A-Y)</math></big><br> | ||
| + | |||
| + | b - Let us express the various energies and matrix elements in terms of the usual <big><math> \beta </math></big> and <big><math> S </math></big> integrals between the X and H orbitals: | ||
| + | |||
| + | <big><math> E_{ind} = \beta S </math></big> | ||
| + | |||
| + | <big><math> S_{12} = 0.5 </math></big> | ||
| + | |||
| + | <big><math> H_{12} = 2 \beta S </math></big> | ||
| + | |||
| + | <big><math> RE = (2 \beta S - 0.5 \beta S )/1.5 = \beta S </math></big> | ||
| + | Since at the transition state geometry <big> <math>\Delta E_{ST} </math>'</big> = <big><math> 4 \beta S </math></big>, we have: | ||
| + | <big> <math>B = 0.25\Delta E_{ST} </math>'</big> | ||
| + | |||
| + | c - Since <big> <math>\Delta E_{ST} </math>' <math> = 2 BDE </math></big>, we may re-express the avoided crossing term as: | ||
| + | |||
| + | <big> <math>B = 0.5BDE </math></big> | ||
| + | |} | ||
=== 2/ Computer exercise === | === 2/ Computer exercise === | ||
| Ligne 253 : | Ligne 375 : | ||
<br> | <br> | ||
<br> | <br> | ||
| − | <big>'''Compute the Energies and Wavefunctions at Reactant and Transition State with Different | + | <big>'''2.1 Computations'''</big> |
| − | + | ||
| + | Compute the Energies and Wavefunctions at Reactant and Transition State with Different Sets of VB Structures | ||
| + | |||
# Write the following VB structure sets, compare and see the difference: | # Write the following VB structure sets, compare and see the difference: | ||
## all structures; | ## all structures; | ||
| Ligne 260 : | Ligne 384 : | ||
## minimal structures for product. | ## minimal structures for product. | ||
# Perform VBSCF and VBCISD calculations for reactant: | # Perform VBSCF and VBCISD calculations for reactant: | ||
| − | ## Perform a VBSCF calculation with "''orbtyp=hao''" and "''boys''"; | + | ## Perform a VBSCF calculation (file name: rs_vbscf) with "''orbtyp=hao''" and "''boys''"; |
| − | ## Perform a VBCISD calculation with VBSCF orbital as initial guess. | + | ## Perform a VBCISD calculation (file name: rs_vbcisd) with VBSCF orbital as initial guess. |
| − | ## Perform VBSCF and VBCISD calculations with minimal structures for reactant and product. | + | ## Perform VBSCF and VBCISD calculations with minimal structures for reactant (file names: rs_*_rs) and product (file names: rs_*_ps). |
| − | # Perform VBSCF and VBCISD calculations for transition state with the same procedure as in step 2. | + | # Perform VBSCF and VBCISD calculations for transition state (file names: ts_*) with the same procedure as in step 2. |
<br> | <br> | ||
| − | <big>'''Analysis: Wavefunctions and Energies'''</big> | + | <big>'''2.2 Analysis: Wavefunctions and Energies'''</big> |
<br> | <br> | ||
# Compute the Barrier height of the <math>\textrm{S}_{\textrm{N}}2</math> reaction at VBSCF and VBCISD levels. See the difference of the barrier heights. | # Compute the Barrier height of the <math>\textrm{S}_{\textrm{N}}2</math> reaction at VBSCF and VBCISD levels. See the difference of the barrier heights. | ||
| Ligne 273 : | Ligne 397 : | ||
<br> | <br> | ||
| − | <big>'''Optional : Compute all points and draw the VBSCDs at VBSCF and VBCISD levels.'''</big> | + | <big>'''2.3 Optional : Compute all points and draw the VBSCDs at VBSCF and VBCISD levels.'''</big> |
| + | <br> | ||
| + | |||
| + | {| class="collapsible collapsed wikitable" | ||
| + | |- | ||
| + | !'''Answer''' | ||
| + | |- | ||
| + | | | ||
| + | a- VB Structures used in the computations | ||
| + | <center>[[File:Structures.png|600px]] </center><br> | ||
| + | <center>Total 8 Structures of The System</center> | ||
| + | <br> | ||
| + | <center>[[File:Reactant_Structures.png|400px]] </center><br> | ||
| + | <center>Structures of The Reactant State</center> | ||
| + | <br> | ||
| + | <center>[[File:Product_Structures.png|400px]] </center><br> | ||
| + | <center>Structures of The Product State</center> | ||
| + | <br> | ||
| + | b- Computational results | ||
| + | <br> | ||
| + | <center> | ||
| + | {| border="1" | ||
| + | |+ Weights of VB Structures of H-H-H Abstract Reaction at Reactant Geometry | ||
| + | |- | ||
| + | ! scope="col" | | ||
| + | ! scope="col" | S1 | ||
| + | ! scope="col" | S2 | ||
| + | ! scope="col" | S3 | ||
| + | ! scope="col" | S4 | ||
| + | ! scope="col" | S5 | ||
| + | ! scope="col" | S6 | ||
| + | ! scope="col" | S7 | ||
| + | ! scope="col" | S8 | ||
| + | |- | ||
| + | ! scope="row" | VBSCF | ||
| + | | 0.803 || 0.003 || 0.096 || 0.001 || 0.095 || 0.000 || 0.000 || 0.000 | ||
| + | |- | ||
| + | ! scope="row" | VBCISD | ||
| + | | 0.770 || 0.005 || 0.110 || 0.003 || 0.111 || 0.000 || 0.000 || 0.001 | ||
| + | |- | ||
| + | |} | ||
| + | </center> | ||
| + | <br> | ||
| + | <center> | ||
| + | {| border="1" | ||
| + | |+ Weights of VB Structures of H-H-H Abstract Reaction at Transition State Geometry | ||
| + | |- | ||
| + | ! scope="col" | | ||
| + | ! scope="col" | S1 | ||
| + | ! scope="col" | S2 | ||
| + | ! scope="col" | S3 | ||
| + | ! scope="col" | S4 | ||
| + | ! scope="col" | S5 | ||
| + | ! scope="col" | S6 | ||
| + | ! scope="col" | S7 | ||
| + | ! scope="col" | S8 | ||
| + | |- | ||
| + | ! scope="row" | VBSCF | ||
| + | | 0.344 || 0.344 || 0.096 || 0.035 || 0.025 || 0.025 || 0.096 || 0.035 | ||
| + | |- | ||
| + | ! scope="row" | VBCISD | ||
| + | | 0.358 || 0.358 || 0.059 || 0.036 || 0.046 || 0.046 || 0.059 || 0.036 | ||
| + | |- | ||
| + | |} | ||
| + | </center> | ||
| + | <br> | ||
| + | <center> | ||
| + | {| border="1" | ||
| + | |+ Energies (a.u.) and Barriers (kcal/mol) of H-H-H Abstract Reaction | ||
| + | ! scope="col" | | ||
| + | ! scope="col" | VBSCF | ||
| + | ! scope="col" | VBCISD | ||
| + | |- | ||
| + | ! scope="row" |Reactant | ||
| + | | -1.64637 || -1.66241 | ||
| + | |- | ||
| + | ! scope="row" | Transition State | ||
| + | | -1.60706 || -1.63827 | ||
| + | |- | ||
| + | ! scope="row" | Barrier | ||
| + | | 24.7 || 15.1 | ||
| + | |- | ||
| + | |} | ||
| + | </center> | ||
| + | <br> | ||
| + | <center> | ||
| + | {| border="1" | ||
| + | |+ Energies(a.u.) and Resonance Energies (''B'', in kcal/mol) of H-H-H Abstract Reaction at Reactant Geometry | ||
| + | ! scope="col" | | ||
| + | ! scope="col" | VBSCF | ||
| + | ! scope="col" | VBCISD | ||
| + | |- | ||
| + | ! scope="row" | All Structures | ||
| + | | -1.64637 || -1.66241 | ||
| + | |- | ||
| + | ! scope="row" | Reactant | ||
| + | | -1.64617 || -1.66208 | ||
| + | |- | ||
| + | ! scope="row" | Product | ||
| + | | -1.40873 || -1.41851 | ||
| + | |- | ||
| + | ! scope="row" | ''B'' | ||
| + | | 0.1 || 0.2 | ||
| + | |- | ||
| + | |} | ||
| + | </center> | ||
| + | <br> | ||
| + | <center> | ||
| + | {| border="1" | ||
| + | |+ Energies (a.u.) and Resonance Energies (''B'', in kcal/mol) of H-H-H Abstract Reaction at Transition State Geometry | ||
| + | ! scope="col" | | ||
| + | ! scope="col" | VBSCF | ||
| + | ! scope="col" | VBCISD | ||
| + | |- | ||
| + | |- | ||
| + | ! scope="row" | All Structures | ||
| + | | -1.60706 || -1.63827 | ||
| + | |- | ||
| + | ! scope="row" | Reactant | ||
| + | | -1.54798 || -1.56655 | ||
| + | |- | ||
| + | ! scope="row" | Product | ||
| + | | -1.54798 || -1.56655 | ||
| + | |- | ||
| + | ! scope="row" | ''B'' | ||
| + | | 37.1 || 45.0 | ||
| + | |- | ||
| + | |} | ||
| + | </center> | ||
| + | <br> | ||
<br> | <br> | ||
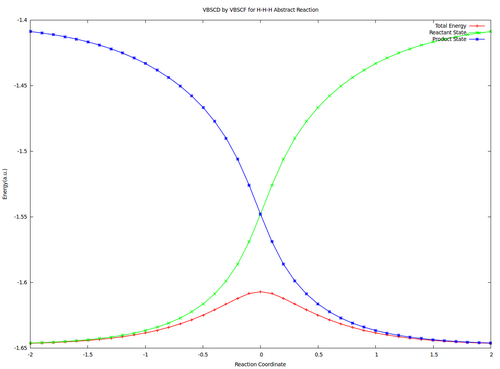
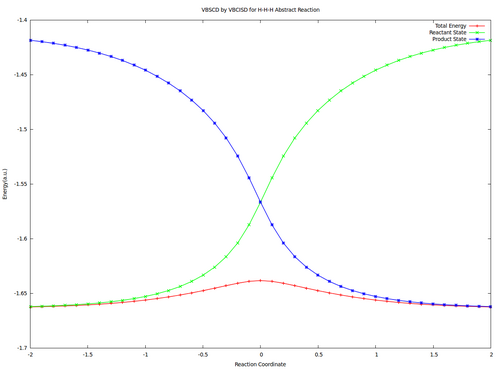
| + | <center><big>Optional : VBSCDs for H-H-H abstract reaction by VBSCF and VBCISD</big></center> | ||
| − | [[ | + | [[File:H3-VBSCD-VBSCF.png|500px]] [[File:H3-VBSCD-VBCISD.png|500px]]<br> |
| + | <big> {{pad|250px}} VBSCF {{pad|420px}} VBCISD </big> | ||
| + | |} | ||
== Exercise 3 (paper exercise) : Conical intersection in H<sub>3</sub><sup>•</sup> radical == | == Exercise 3 (paper exercise) : Conical intersection in H<sub>3</sub><sup>•</sup> radical == | ||
| Ligne 300 : | Ligne 556 : | ||
<big><math> <D|H|D'>=<|...i\overline{j}...||H||...\overline{i}j...|>= -2 \beta_{ij} S_{ij}</math></big> (for <big><math>D</math>, <math>D'</math></big> differing by spin inversion of two spin-orbitals) | <big><math> <D|H|D'>=<|...i\overline{j}...||H||...\overline{i}j...|>= -2 \beta_{ij} S_{ij}</math></big> (for <big><math>D</math>, <math>D'</math></big> differing by spin inversion of two spin-orbitals) | ||
| − | + | {| class="collapsible collapsed wikitable" | |
| + | |- | ||
| + | !'''Answer''' | ||
| + | |- | ||
| + | | | ||
| + | <math> R = \frac{1}{\sqrt{2}} (|ab\overline{c}|-|a\overline{b}c|) </math><br> | ||
| + | <math> P = \frac{1}{\sqrt{2}} (|a\overline{b}c|-|\overline{a}bc|) </math><br> | ||
| + | |||
| + | 1 - Using the thumb rules | ||
| + | |||
| + | <big><math> <D|H|D>= -2 \sum_{i<j}^{ } \beta_{ij} S_{ij} </math>(if orbitals i and j have parallel spins) </big> | ||
| + | <big><math> <D|H|D'>=<|...i\overline{j}...||H||...\overline{i}j...|>= -2 \beta_{ij} S_{ij} </math>(for D, D' differing by spin inversion of two spin-orbitals</big> | ||
| + | |||
| + | It comes,<br> | ||
| + | <big><math> <R|H|R>= \frac{1}{2} (4\beta_{bc}S_{bc} -2\beta_{ab}S_{ab} -2\beta_{ac}S_{ac})</math></big><br> | ||
| + | <big><math> <P|H|P>= \frac{1}{2} (4\beta_{ab}S_{ab} -2\beta_{bc}S_{bc} -2\beta_{ac}S_{ac})</math></big><br> | ||
| + | |||
| + | <big><math> <R|H|P>= \frac{1}{2} (4\beta_{ac}S_{ac} -2\beta_{ab}S_{ab} -2\beta_{bc}S_{bc})</math></big><br> | ||
| + | |||
| + | |||
| + | 2 - If <math>\theta </math> > 60° ,<big> <math>\beta_{ac}S_{ac} < \beta_{ab}S_{ab}=\beta_{bc}S_{bc} </math></big> Hence <big><math><R|H|P> > 0 </math></big> (<math>\beta_{ij} < 0</math>) <br> | ||
| + | |||
| + | Finally, ground state is out of phase and the excited state is in phase. | ||
| + | |||
| + | It comes:<br> | ||
| + | :<big><math> \Psi^{\neq} = R-P</math></big><br> | ||
| + | :<big><math> \Psi^{\star} = R+P</math></big><br> | ||
| + | |||
| + | Expanding on the determinants | ||
| + | :<big><math> \Psi^{\neq} = 1/\sqrt{2} (|ab\overline{c}|+|\overline{a}bc|-2|a\overline{b}c|)</math></big><br> | ||
| + | :<big><math> \Psi^{\star} =1/\sqrt{2} (|ab\overline{c}|-|\overline{a}bc|)</math></big><br> | ||
| + | |||
| + | <math> \Psi^{\star} </math>, wave function of the excited state represent a bonding between hydrogens a and c.<br> | ||
| + | |||
| + | 3- Using geometric considerations | ||
| + | * If <math>\theta </math> = 180° ,<big> <math>\beta_{ac}S_{ac}=0</math></big> and <big><math> <R|H|P>=-(\beta_{ab}S_{ab} +\beta_{bc}S_{bc})</math> </big><br> | ||
| + | |||
| + | * If <math>\theta </math> = 60° ,<big> <math>\beta_{ac}S_{ac}=\beta_{ab}S_{ab}=\beta_{bc}S_{bc}=\beta S</math></big> and <big><math> <R|H|P>=0</math></big> <br> | ||
| + | |||
| + | 4- If <math>\theta </math> = 60°, <big><math> <R|H|P>=0 = <R|H|R>= <P|H|P></math></big> <br> | ||
| + | Hence R and P are degenerated eigenfunction of the CI Hamiltonian : <math>E_R=E_P=0</math><br> | ||
| + | Their linear combination <math> \Psi^{\neq} </math> and <math>\Psi^{\star} </math> are also degenerated, with the same value. | ||
| + | |||
| + | 5- Allyl radical <math>\pi</math> system is isoelectronic to the <math>H_3</math> radical case. | ||
| + | R corresponds to the covalent right coupling (radical on the left carbone atom). P to the covalent right coupling. | ||
| + | * <math> \Psi^{\neq} </math> corresponds to the resonance (mesomery) between these 2 bonding schemes. | ||
| + | * <math>\Psi^{\star} </math> to the "through space" (a,c) electronic coupling ("long bond", the radical is centered on the middle atom). | ||
| + | |||
| + | The geometrical distortion to make the two states degenerated in allyl radical would be a rotation of the two CH2 end groups: by such a rotation, the resonance structures are destabilized (pi bonds break), and the a-c coupling is re-inforced (sigma bond forms). | ||
| + | |||
| + | The end product obtained from the first excited state of allyl radical is a cyclopropyl radical (formation of the a-c bond). | ||
| + | |||
| + | |||
| + | |} | ||
|} | |} | ||
| − | + | ||
| + | |||
| + | |||
| + | |||
| + | ---- | ||
Dernière version du 18 janvier 2013 à 15:56
Valence Bond State correlation diagrams
Exercise 1 : Computation of state correlation Diagrams for a 3 centers / 4 electrons system
In this exercise the <math>\textrm{S}_{\textrm{N}}2</math> reaction Cl<math>{}^{-}</math> + CH<math>{}_3</math>Cl -> ClCH<math>{}_3</math> + Cl<math>{}^{-}</math> will be studied in both vacuum and solution. Valence Bond State Correlation Diagrams (VBSCD) will be constructed at <math>\pi</math>-D-BOVB level. There are two parts in this exercise: basic part and optional part. The basic part is performed with MCP-DZP basis set in which the inner orbitals in Cl and C are described with MCP pseudo potential. The optional part is performed with 6-31+G* basis set, using the general specification for the xmvb input (expert users). Only reactant and transition state will be computed in this exercise, which is sufficient to build the VBSCD diagrams.
| Note:How to perform a VBPCM calculation |
|---|
|
A VBPCM calculation is performed in the similar way as the VB calculations in vacuum. One should prepare a GAMESS input file with solvent assigned such as: The details of PCM calculation in the GAMESS can be found in GAMESS manual. Keyword "VBTYP=XMVB" in CONTRL section is also essential. After the GAMESS input file is prepared, an XMI file with keyword "VBPCM" should be prepared with the same file name as GAMESS input file. In the current XMVB package, VBSCF/PCM and BOVB/PCM calculations are both supported. |
| Basic part | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
1. Compute the Energies and Wavefunctions at Reactant and Transition State with Different Sets of VB StructuresWe advise you to create the first xmvb input file (.xmi file) for this study starting from a .xmi file taken from tutorial1 as a template. Alternatively, you may copy the input file corresponding for the first calculation of this study (L-VBSCF on reactant state geometry in vacuum) from the answer folder of this exercise (cp answer/rs_vac_vbscf.xmi .), run directly the first calculation, inspect input/outputs, and then use this .xmi file as a template for the following calculations.
2. Analysis: Wavefunctions and Energies
|