Différences entre les versions de « VBTutorial1 »
| Ligne 76 : | Ligne 76 : | ||
== Exercise 4 : F<math>{}_2</math> molecule and charge-shift resonance energy == | == Exercise 4 : F<math>{}_2</math> molecule and charge-shift resonance energy == | ||
| + | |||
| + | <big><big><center><font color=orange> '''***** INPUT FILES TO BE FINALIZED *****''' </font></center></big></big> | ||
| + | |||
# Compute a VBSCF wave function for the F<math>{}_2</math> molecule, with inactive orbitals localized on only one of the fluorine atoms ; | # Compute a VBSCF wave function for the F<math>{}_2</math> molecule, with inactive orbitals localized on only one of the fluorine atoms ; | ||
## first the ''frgtyp=sao'' specification and automatic guess (''guess=auto'') ; | ## first the ''frgtyp=sao'' specification and automatic guess (''guess=auto'') ; | ||
Version du 23 juin 2012 à 09:37
How to modify this page :
- first : log in (top right of this page) ;
- click on [edit] (far right) to edit a section of the page ;
- write your text directly in the wiki page, and click on the "Save page" button (bottom left) to save your modifications
Pictures : how to insert a picture in your text
See also this page for an introduction to the basics of the wiki syntax
Remarks
Exercises
Exercise 1 (paper exercise) : The lone pairs of H2O
(for further reading, see S. Shaik and P.C. Hiberty, "The Chemist's Guide to VB theory", Wiley, Hoboken, New Jersey, 2008, pp. 107-109)
This exercise aims at comparing two descriptions of the lone pairs of H<math>{}_2</math>O : (i) the MO description in terms of non-equivalent canonical MOs and (ii) the « rabbit-ear » VB description in terms of two equivalent hybrid orbitals.

- Focusing on the lone pairs only, write the four-electron single-determinants <math>\Psi_{\textrm{MO}} </math> and <math>\Psi_{\textrm{VB}} </math> .
- Expand <math>\Psi_{\textrm{VB}} </math> into elementary determinants containing only <math>n</math> and <math>p</math> orbitals, eliminate determinants having two identical spinorbitals, and show the equivalence between <math>\Psi_{\textrm{VB}}</math> and <math>\Psi_{\textrm{MO}}</math>.
- We now remove one electron from H<math>{}_2</math>O. Write the two possible VB structures <math>\Phi_1</math> and <math>\Phi_2</math> in the VB framework.
- The two ionized states are the symmetry-adapted combinations
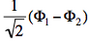 and
and 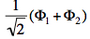 . From the sign of the hamiltonian matrix element <math>\langle \Phi_1 \vert \hat{H} \vert \Phi_2 \rangle</math>, give the energy ordering of the two ionized states.
. From the sign of the hamiltonian matrix element <math>\langle \Phi_1 \vert \hat{H} \vert \Phi_2 \rangle</math>, give the energy ordering of the two ionized states. - By expanding the two ionized states into elementary determinants (dropping the normalization constants), show that they are equivalent, respectively, to the MO configurations <math>\vert nn\bar{p}\vert</math> and <math>\vert pp\bar{n}\vert</math>.
Appendix
Hamiltonian matrix element between determinants differing by one spin-orbital :
Don’t forget to put the orbitals of the two determinants in maximal correspondence before applying the rule.
Exercise 2 : Starting up with the H<math>{}_2</math> molecule
Two Gamess and XMVB input files for the H<math>{}_2</math> molecule are provided in the Exercise folder on the tutorial machines :
- the file h2-atom.xmi input uses the fragment specification in terms of atoms (frgtyp=atom) ;
- the file h2-sao.xmi input uses the fragment specification in terms of symmetry-adapted orbitals (frgtyp=sao).
There are VBSCF calculations with the 6-31G(d,p) basis set. Just inspect these inputs, run the gamess-xmvb program (using : vbrun h2-atom and : vbrun h2-sao, and analyze the outputs.
Then these input files could serve you as templates for the next exercises.
Exercise 3 : HF molecule : weights and bond energy
- Compute a VBSCF three structure wave function for the HF molecule, using the frgtyp=sao specification and automatic guess (guess=auto). Which structure(s) should be kept in further BOVB calculations ?
- Using VBSCF orbitals as guess orbitals :
- Compute a L-BOVB wave function on a selected subset of structures ;
- Compute a VBCISD wave function for the multi-structure wave function
- Compare structure weights at the VBSCF, L-BOVB and VBCI levels
- Compute bond energies at the L-BOVB and VBCISD levels.
Exercise 4 : F<math>{}_2</math> molecule and charge-shift resonance energy
- Compute a VBSCF wave function for the F<math>{}_2</math> molecule, with inactive orbitals localized on only one of the fluorine atoms ;
- first the frgtyp=sao specification and automatic guess (guess=auto) ;
- second the frgtyp=atom specification and providing HF MOs as guess orbital through an extra $Gus section in the xmvb input
- BOVB level :
- Compute a L-BOVB wave function using VBSCF orbitals as guess orbitals ;
- Starting from the previous solution, compute a πD-BOVB solution, by allowing only the π lone pairs to delocalize onto the two atoms. Compare total energy with the previous level.
- Calculate the charge-shift resonance energy for the F<math>{}_2</math> molecule. Compare it with the bond energy (Experimental : ~40 kcal/mol).
Exercice 5 : Solvent effect on C(Me)<math>{}_3</math>-Cl weights
- C(Me)<math>{}_3</math>-Cl at equilibrium geometry :
- Compute a VBSCF wave function using frgtyp=atom and a $Gus section to specify guess orbitals. The active electron pair will be the C-Cl bond, and all inactive orbitals should be localized either on Cl or on the C(Me3) fragment. Which structures should be kept in further BOVB calculations ?
- Compute a L-BOVB wave function.
- Compute a D-BOVB wave function, by freezing the active orbitals, and delocalizing all inactive orbitals onto the whole molecule.
- Starting from guess orbitals obtained at equilibrium geometry, redo the D-BOVB calculation for the large inter fragment distance. How does the weights of the different structures evolve when the molecule is stretched ?
- Redo the D-BOVB calculations at equilibrium geometry and large distance using VBPCM for water. How does the weights change with solvation effects ?