Training for the NCI workshop
<<< Topological Approaches to Intermolecular Interactions workshop main page
| This is a special session organised by R Chaudret
PLACES ARE NOT ENSURED BEYOND 12 SEATS; TO BE DETERMINED BY ROOM AND TIME AVAILABILITY |
| Practical trainings will be organized on the prior morning and/or afternoon to the workshop.
The following training activities are envisaged:
|
PARTICIPANTS TO THE TRAINING SESSION: Please fill the following doodle web page:
http://doodle.com/exzhkiegergybmd9
Speakers of the training session, please add your abstract below:
- first : log in;
- click on "edit", on the righthand corner above the the line below "Your name"
- copy the example given (also available >>> here) and paste it at the end of this page
- fill in the fields with your data
- >>> How to insert a picture in your abstract
- Don't forget to send us an email with the type of contribution you would like to make
NCI (basics) : first steps into NCIPLOT and VMD
Julia Contreras-Garcia
NCI (Non-Covalent Interactions) is a visualization index based on the density and its derivatives. It enables identification of non-covalent interactions. It is based on the peaks that appear in the reduced density gradient (RDG) at low densities.
There is a crucial change in the RDG at the critical points in between molecules due to the annihilation of the density gradient at these points. When we plot the RDG as a function of the density across a molecule, we see that the main difference between the monomer (Figure 1a) and dimer (Figure 1b) cases is the appearance of steep peaks at low density.
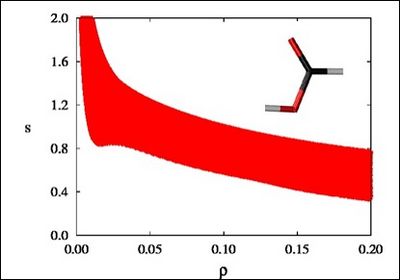
Figure 1a |
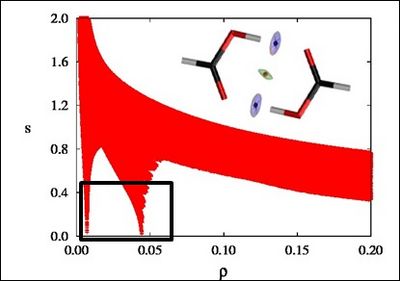
Figure 1b |
When we search for the points in 3D space giving rise to these peaks, non covalent regions clearly appear in the (supra)molecular complex (insets in Figure 1a and Figure 1b).
During this training, we will review how to carry out NCI calculations, both from promolecular and SCF calculations. We will also go into the VMD script, so that everybody knows how to visualize their output!
ELF (basics) : first steps into TOPMOD and topological analysis
Benoit de Courcy
Coupling ELF and NCI
Robin Chaudret
In this final training of the morning session we will emphase on the complementarity of ELF and NCI approaches and particularly to study reaction mechanisms. Indeed, since ELF focuses on strong density area and nci on weak density ones, these approaches allow to follow the modification of the system at any step of the reaction. During the training we will specifically learn how to understand an ELF/NCI picture and to use different scripts to generate automatically ELF/NCI short movie.
Averaged NCI: using NCI for dynamic systems
Robin Chaudret
To apply the NCI analysis to fluctuating environments as in the solution phase, we developed a new averaged noncovalent interaction (i.e., aNCI) index along with a fluctuation index to characterize the magnitude of interactions and fluctuations. We implemented them into the NCIplot programm and tested them on several solute/solvant but also substrate/enzyme systems. During the training session we will rapidly review the theory of aNCI and how and why we decided to go from NCI to aNCI. Then we will do practical exercices to learn the usage of the specific NCIplot keywords going along with an aNCI calculation. We will also review several scripts that allow to create aNCI input.
NCI for solids
Alberto Otero-de-la-Roza
NCI from experimental densities
Gabriele Saleh
Department of Chemistry, Università degli Studi di Milano, via C.Golgi 19, 20159 Milano Italy
NCImilano: a code to apply non-covalent interactions descriptor to experimental and theoretical electron density distribution
The code “NCImilano” [1] is designed to apply the RDG-based NCI descriptor introduced by Johnson et al. [2] to Electron Density (ED) distribution obtained either from theoretical calculation (on both isolated molecules and crystals) or from X-ray diffraction experiments [3,4]. More specifically, it can read the output files of GAUSSIAN 03/09, TOPOND (which is interfaced to CRYSTAL 98/06/09) and XD2006. When files in XD2006 format are given in input to “NCImilano”, output files in the same format are produced, so that they can be read from the graphical routine of XD. Besides calculating Reduced Density Gradient (RDG) and ED*sign(λ2) (where λ2 is the second greatest eigenvalue of the ED Hessian matrix), i.e. the quantities needed to apply the NCI descriptor in its original formulation, this program can also produce grid files of total energy density along with its potential and kinetic contribution. The latter quantities can be evaluated either exactly from the wavefunction or, in the case of experimentally-derived ED (i.e. when the wavefunction is not available), by exploiting the approximate functional introduced by Abramov [5]. In addition, NCImilano compute the volume of RDG isosurfaces and the integral of several quantities over the space enclosed into such isosurfaces. In this talk the main features of the code NCImilano will be presented, with special emphasis on the calculation of properties from experimentally-derived ED.
References [1] G. Saleh, C. Gatti, L. Lo Presti, D. Ceresoli. Submitted to J. Appl. Cryst. on 15th March 2013 [2] E. R. Johnson, et al. (2010) J. Am. Chem. Soc. 132, 6498–6506 [3] G. Saleh, C. Gatti, L. Lo Presti, J. Contreras-Garcìa (2012) Chem. Eur. J. 18, 15523-15536 [4] G. Saleh, C. Gatti, L. Lo Presti, L. (2012) Comput. Theor. Chem. 998,148–163 [5] Y. A. Abramov (1997) Acta Cryst. A53, 264-272
Maximum Probability Domains (MPDs)
Benoit Braida